If Phylogenetic Sense Be Something Biologists Wish
- Rohini Subrahmanyam

- Aug 8, 2023
- 11 min read
Updated: Feb 19
Nearly two decades in, researchers are still debating which creature is the closest living relative to all animals. Are new methods getting them any closer?
The affinities of all the beings of the same class have sometimes been represented by a great tree. I believe this simile largely speaks the truth. — Charles Darwin
Ever since Darwin posited that all species had a common ancestor, scientists have been trying to map out the evolutionary tree and fill it with different organisms based on when and how they must have evolved on this earth. The evolution of animals is something a lot of scientists care about because we humans ourselves belong to the animal kingdom: How did we get here? What makes us human? Where do we go from here? Sadly, as we don’t have a time machine, we have to rely on scientific estimates for this.
Let's Dig Deep Together.
Subscribe to The Xylom's free weekly newsletter for more in-depth reporting
For many years before the turn of the century, it was assumed that sponges belonged to the oldest lineage that split off from the last common ancestor, or the sister-group of all animals. A 2021 Nature study has dated some of the oldest sponge fossils to nearly 890 million years ago. But as these creatures have mostly soft gelatinous bodies, and also lived in water (perhaps not in a pineapple under the sea though), the fossil record can be spotty.
“For a long time, everyone had worked under this paradigm that the earliest animals were the simplest animals,” said Gonzalo Giribet, a professor in the Department of Organismic and Evolutionary Biology and the Director of the Museum of Comparative Zoology at Harvard University.
“For a long time, everyone had worked under this paradigm that the earliest animals were the simplest animals.”
Animals without any organ systems, without even any tissues, seemed like the most likely candidate for being part of the oldest animal lineage. And so sponges, being relatively simple aquatic animals, made a lot of sense to be at the base of the animal tree, the sister group of all other animals. There were some animals even simpler than sponges, placozoans, which had even lesser types of cells. Scientists even considered placing placozoans earlier in the evolutionary tree than sponges, very briefly. But phylogenetic studies, or studying evolutionary history using genetic data, quickly ruled them out.
Little did they know; the battle for the oldest animal lineage hadn’t even begun yet. A different tiny, marine invertebrate was about to enter the arena.
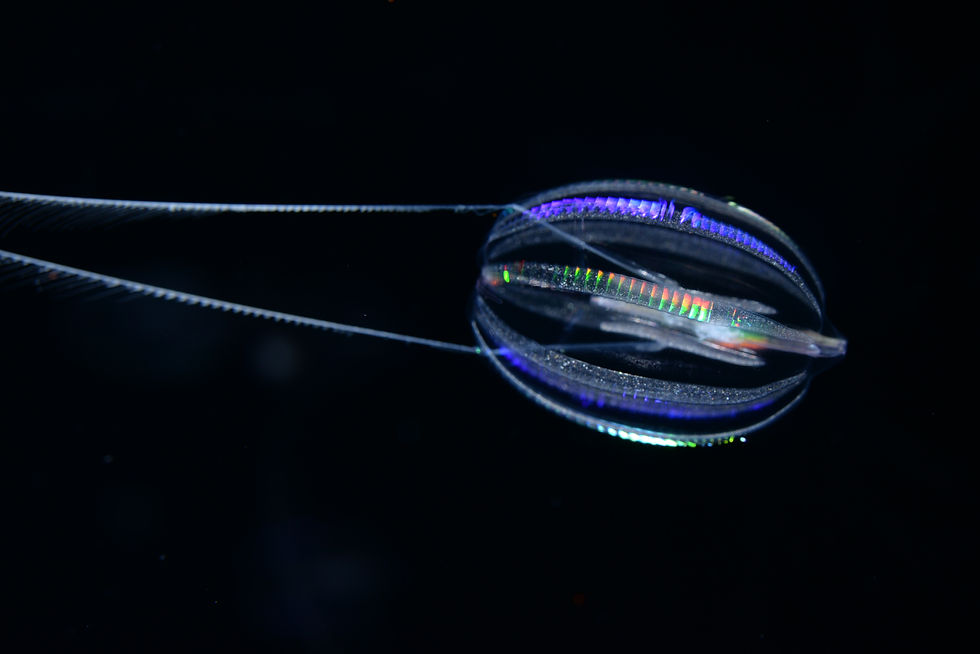
“Ctenophores, on the other hand, are more complex. They have a nervous system, they have muscle cells,” said Giribet.
Ctenophores (pronounced Teen-nophores), better known as comb jellies, are also simple marine invertebrates. But they are slightly more complex than sponges, containing some primitive organ systems inside them. More than a decade ago, a team led by Casey Dunn, now a Professor of Ecology and Evolutionary Biology at Yale University, didn’t have access to many sequenced genomes, but what they had was an old-fashioned style of RNA sequencing to look at what the genes could make.
ADVERTISEMENT
“We did not have an axe to grind, we weren’t looking for a particular result; we wanted to do things the right way,” Mark Martindale, Director of the Whitney Laboratory for Marine Bioscience at the University of Florida who collaborated with Dunn and Giribet on the study, insisted.
“We did not have an axe to grind, we weren’t looking for a particular result; we wanted to do things the right way,.”
Using this readout from all the animals they could get their hands on, the scientists decided to draw out the entire evolutionary tree of animals. “We did this analysis completely naïve, we put everything in, and we found ctenophores branching before sponges. That was really shocking to us, we did a lot of analysis and tested a lot of things, but we got this result,” said Giribet.
That result, published in Nature in 2008, didn’t just shock them but shocked the entire field, and kickstarted a huge debate between evolutionary biologists over which animal branched off first from the last common ancestor of all animals.
Sponges had been the unrivaled oldest animal lineage and the sister group to all other animals for so many years, all our textbooks had them. Suddenly, these little shimmering jelly-like creatures, with their iridescent combs and more importantly a nerve net that controls their muscle cells, had beaten back with their microscopic cilia the claims to evolutionary bragging rights. From an organism shackled to the ocean floor for the rest of its adult life, absorbent and porous is he? The idea that something more complex than the simple sponge had split off first from the common ancestor was especially irksome to a lot of scientists.
“People couldn’t wrap their heads around that. Simple animals have to have simple genomes, and there has to be some sort of relation between genomic and morphological complexity. When we showed this was not true… it completely knocked out this simple-minded, human idea of how evolution has to work.”
In 2007, a year before their controversial paper, Martindale, Daniel Rokhsar, a Professor of Genetics, Genomics, Evolution, and Development at UC Berkeley, and others published the sequenced genome of a little sea anemone, one of the first cnidarians or jelly-fish type animals to be sequenced. These creatures are also marine but are known to be relatively closer in their evolutionary origins to organisms with bilateral symmetry, or biltarians, like vertebrates and humans. But they are still not very complex beings in terms of morphology; they have very few organ systems and a diffuse nerve net like in the comb jellies.
Much to everyone’s surprise, they showed that sea anemones had more genes in common with human beings than even fruit flies or worms, which are closer relatives to humans. This means that more “complex” animals which are more closely related to humans, have actually lost genes that were in the common ancestor of “simple” sea anemones and humans.
“People couldn’t wrap their heads around that. Simple animals have to have simple genomes, and there has to be some sort of relation between genomic and morphological complexity. When we showed this was not true… it completely knocked out this simple-minded, human idea of how evolution has to work,” explained Martindale.
“Thinking that everything goes from simple to complex hasn’t worked many times in the Tree of Life,” said Giribet.
After the 2008 paper by Dunn, Martindale, and Giribet, this tiny marine organism, the comb jelly, became the center of a huge, heated debate between scientists for the next decade or so. Multiple papers followed, each paper analyzing and comparing the genomic sequences of ctenophores and sponges and different animals. Some papers found ctenophores at the base of the tree and some papers reported sponges to be at the base. If we really come down to it, a major limiting factor seemed to be the lack of a perfectly reliable method to analyze the sequence data, and different groups seemed to get different results based on their method.
“It’s a very heated debate. Even for those of us who are involved, it’s a real mire in terms of methodology and it comes to very minute detail,” said Anthony Redmond, a Research Fellow at the Smurfit Institute of Genetics of Trinity College Dublin in Dublin, Ireland.
ADVERTISEMENT
Analyzing these genomic sequences is not easy. Twenty standard amino acids are formed by specific combinations of three consecutive bases, and proteins are formed by the 3-D arrangement of various amino acids. Bases are notorious for changing back and forth, accordingly changing the amino acid and protein as well — at least most of the time because some amino acids can be coded with multiple combinations of bases, and changes to amino acids could mean everything between “not much” and a protein with a completely different function. The longer back in time you go, the more genes could change, but also the more likely these changes could have reversed along the way, increasing the difficulty of retracing the exact chronological order of all these changes. Another issue with genetic sequences is that distantly related species may converge on similar sequences just by chance.
“In all of the studies up to now, the best efforts to model the amino acid evolution well, in a realistic fashion, point towards sponges. However, we know that all of the models we have till now don’t do particularly well,” said Redmond.
According to Redmond, when scientists used models to correct for the issues faced due to amino acids changing multiple times, or convergent sequences in distant species, the answer would point towards sponges at the base of the tree. (Redmond himself was an author of a 2021 Nature paper that concluded sponges were the sister group to all other animals.)
If we were to go by British statistician George Box’s words, “all models are wrong, but some are useful,” then in Redmond’s opinion, the “sponge-first” models were decidedly more useful.
That is, until a new study burst into the scene this May.

Oleg Simakov, an Assistant Professor of Molecular Evolution and Development at the University of Vienna, has been working on organisms that have interesting and important positions on the phylogenetic tree for many years.
“The bottleneck, back in the days, was that we didn’t have chromosomal scale genomes. Genomic information was limited to short pieces of genomic sequences,” said Simakov.
In animals like fruit flies or mice, we have known for much longer how genes are arranged together on a chromosome, but for animals like comb jellies, hydra, or sponges, this was not possible until very recently. With newer technology, it has been easier to look at a chromosome-level organization of genes, to not just look at sequences within a gene, but to study how different genes are linked. Genetic sequences are made up of single bases, which are known to change many times over the course of evolution, but different genes are linked over large portions of the chromosome. The odds of huge pieces of DNA changing around multiple times during evolution are much lesser.
ADVERTISEMENT
Darrin Schultz, a postdoctoral researcher working with Simakov, was interested in the evolution of 3-D genome structures in early animals. As part of his work, he had to look at many genes on many chromosomes, and while doing that he found evidence of conserved synteny in certain animals. This means that he saw that large portions of the DNA, or whole regions of the chromosomes, were similar between certain animals.
“We realized that the patterns had implications for understanding evolutionary history and could help us polarize the relationship between sponges and ctenophores and all animals,” said Schultz. “It was kind of like a chain reaction that we stumbled upon.”
When he looked at the chromosomes of sponges and all other animals, he found that the chromosomes had fused, and genes had gotten inter-mixed between the two chromosomes. But when he looked at chromosomes in ctenophores, and in unicellular organisms like choanoflagellates (the closest relatives to animals) the chromosomes and the genes were separate and not inter-mixed. This inter-mixing requires huge portions of the DNA to have moved around.
Simakov likened it to adding milk to coffee — it could only have happened once, and it is irreversible. It appeared that based on these events, ctenophores did branch off first from the last common ancestor to choanoflagellates and animals and never underwent inter-mixing events. Sponges, on the other hand, underwent these inter-mixing events, and so did all other animals, making them more closely related. Based on this new data published in Nature, using their new method of looking at chromosomal synteny, the researchers have tipped the scales more towards the ctenophore-sister hypothesis.
“It was kind of like a chain reaction that we stumbled upon.”
“Ctenophores themselves are very fast-evolving species, they are very enigmatic. Their chromosomes look a bit weird, their biology is super interesting and weird. A lot of things have gone on in those animals since they split off,” said Simakov. “It was always a problem in previous phylogenetic studies, is it because of a very fast evolutionary rate that they were at the base of the phylogenetic tree or are they truly early branching animals?”
When asked about the finding by Simakov, Schultz, and their colleagues, Martindale praises the new method for pushing the envelope on phylogenetic analysis. “The recent paper was a real step forward because it gets around some of the problems people are having on agreeing what is the best program to do the analysis with.” This paper is changing the minds of even some of the most ardent believers of the sponge-sister hypothesis. “This is definitely, in my opinion, the most I have been swayed towards comb jellies,” said Redmond.
ADVERTISEMENT
He is however hesitant to call this the smoking gun that puts an end to the debate. “The debate has gone on for so long that I think… there is going to be a post-mortem type situation where there are going to be more papers to try to figure out why we were so wrong, why the debate existed before with the traditional methods, or if the new method is as fool-proof as it seems to be at face value now. Any time you introduce a new method, even if it seems intuitive, we need to understand the nuts and bolts of what’s going on.”
“...there is going to be a post-mortem type situation where there are going to be more papers to try to figure out why we were so wrong, why the debate existed before with the traditional methods, or if the new method is as fool-proof as it seems to be at face value.”
Davide Pisani, a Professor of Phylogenomics at the University of Bristol, who published a paper on Current Biology in 2017 with data supporting the sponge-sister hypothesis, is cautious about the results using the new method. “We are very far from a conclusion of this debate,” he said, mentioning how we need to look at the problem using all the known methods and find a resolution.
“Ultimately, we need to achieve some sort of congruence between fusions, [the presence or absence] of protein-coding genes, and sequence data. Every data type has independent strengths, weaknesses, and idiosyncrasies; perfect data does not exist.”

Will this debate between scientists over the oldest animal lineage ever really end? It is impossible to say, right now, because, like any good science problem, there are always multiple hypotheses that are all falsifiable (one that could be logically contradicted through an empirical test but just not yet.)
If we are to believe the new evidence pointing towards comb jellies as the oldest animal lineage, another topic that is heavily debated among scientists comes to the fore is, how did the nervous system evolve in these creatures? Humans can’t see or hear or feel or think or move or respond to the world if not for all the neurons that populate our bodies. Ctenophores have a diffuse nerve net and other kinds of neurons, but sponges don’t really have any nervous system to speak of. Did sponges, if they diverged later, lose their nerves? Or did ctenophores evolve a nervous system all on their own, independently? The questions are fascinating and endless, and scientists, as always, have multiple hypotheses to test.
ADVERTISEMENT
Schultz is excited about the results of his paper and the implications it has for the field. “Resolving the ctenophore-sponge debate is important because it helps us understand animal evolution a little better. Knowing the topology of animal evolution, knowing that ctenophores have been on their own lineage for a long amount of time could help us understand the evolution of neurons.”
We don’t know if this new finding that shows strong evidence towards ctenophores being the oldest animal lineage truly settles the debate and pushes sponges out of the race — headlines like “Evolution row ends as scientists declare sponges to be sister of all other animals” now look premature in retrospect. But just like Spongebob tries (and fails) at jellyfishing, we may try (and fail) at capturing the complete phylogenetic tree of the living world. But that’s okay. We learn a bit more about the world and ourselves in the process, and we try again. That’s being human.






